PTMVision
Interactive Exploration of Post Translational Modifications
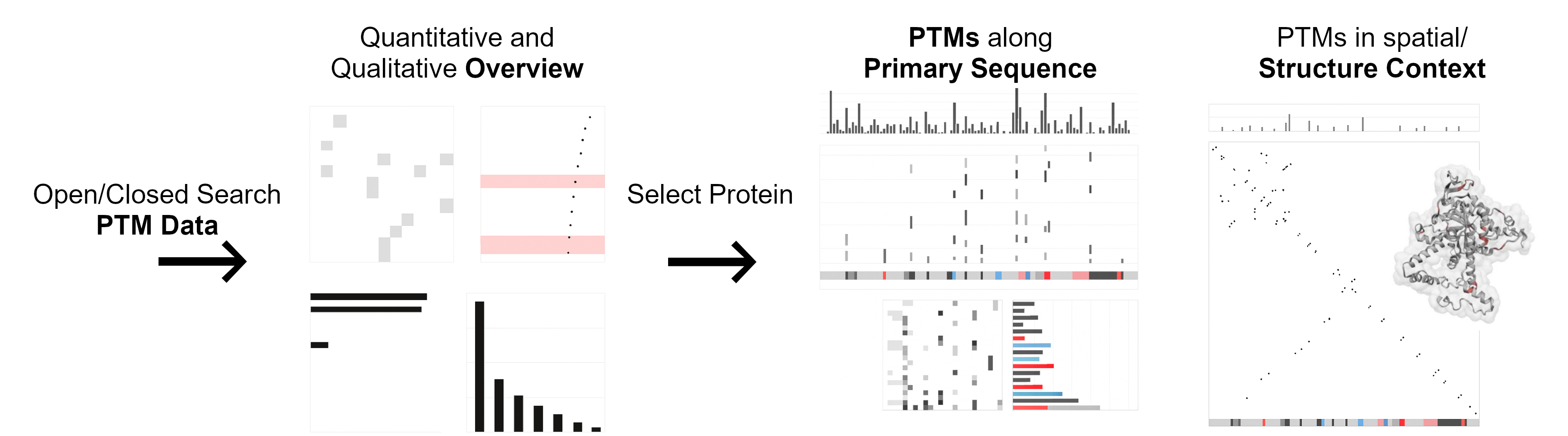
PTMVision supports the upload of PTM data from various proteomics search engines, the mzIdentML standard or manually compiled CSV files. Receive a quantitative and qualitative assessment of the PTMs across all proteins in your data; and get an overview of all available proteins from your data, complemented with associated PTMs. For selected proteins, investigate detailed information on the occurrence of PTMs along the primary protein sequence and in a spatial structural context.
News
08/15/2025
- Add support for PeptideAtlas data.
Background
Understanding the intricate landscape of PTMs is crucial for unraveling the complex mechanisms underlying cellular processes and diseases. PTMVision (Interactive Visualization of Post Translational Modifications) is an interactive web platform for effortless exploratory visual analysis of post translational modifications (PTMs) obtained from mass spectrometry (MS) data using search engines such as ionbot, MaxQuant, MSFragger, Spectronaut, or Sage. The application is designed in close collaboration with proteomics experts and was initiated as an entry to the Bio+MedVis Challenge at the IEEE VIS conference in 2022. PTMVision is accessible to laboratory researchers without requiring coding skills.
Features
- Variety of overview and summary visualizations that help identify patterns and trends in PTM data.
- Visualizations are dynamically linked and support features to highlight PTMs of interest.
- Integration of protein structure and annotation data from UniProt provide multidimensional view. This improves the interpretability of the data significantly.
- Unique emphasis on interactions of residues in close contact within a single protein and associated modifications. This allows to study the dynamic interplay between PTMs and their putative effects on protein folding and function.
- Responsive and intuitive interface that allows users to effortlessly navigate the website and through their data.
- Supports various input formats and an easy export of session data to continue analysis without repeated processing.
Limitations
- We recommend using PTMVision in Chrome or Firefox for optimal performance. Safari has issues when visualising larger datasets.
- Note that PTMVision serves solely to visualize the content provided in the input and does not conduct any further quality control. The localization of modifications still remains a challenging task and may yield imprecise results.
- PTMVision will be continuously updated and improved, including the addition of filters for localisation scores as soon as they will be reported by search engines.
- PTMVision currently limits the maximal request size with respect to the zlib compressed content of the PTM Data file to a maximum of 50 MB.
If you use this software, please cite it as below:
PTMVision: An Interactive Visualization Webserver for Post-translational Modifications of Proteins
Simon Hackl, Caroline Jachmann, Mathias Witte Paz, Theresa Anisja Harbig, Lennart Martens, and Kay Nieselt
Journal of Proteome Research 2025 24 (2), 919-928. DOI:
10.1021/acs.jproteome.4c00679
Start Session
Upload data, re-upload a session or browse and start an example session. Once complete, session data can be downloaded with Download Session Data in the header menu and panels and will be filled. Click symbols for help.
PTMVision sessions are stored for 24 hours on our server and will be deleted afterwards (existing session data is automatically re-initialized).
Upload post-translational modification data from open or closed search to start a new session:
Mandatory Input
Format
Mandatory PTM Data
Optional Exclude UniMod
Classes
Optional Adjust Mass
Shift Tolerance
(Or) Re-upload a PTMVision session:
Mandatory PTMVision
Session
(Or)
Example sessions to explore the features of PTMVision can be started from the table below by clicking the individual Start buttons - the PTM Data files used for the sessions can be downloaded from the PTMVision Zenodo record at https://zenodo.org/records/13270569.
| Search Engine or File Format | Version | Search Type | Source Data | Note | |
|---|---|---|---|---|---|
| ionbot | v0.11.3 |
Open | PRIDE project PXD000498, LC-MS/MS run of an unenriched Escherichia coli sample. | UniMod classes Artefact and Chemical derivative excluded (via PTMVision). | |
| MSFragger | v4.0 |
Open | PRIDE project PXD007740, LC-MS/MS run of a human colorectal cancer sample enriched for phosphorylated peptides using Ti4+-IMAC. | Input file filtered for modified peptides. UniMod classes Artefact and Chemical derivative excluded (via PTMVision). | |
| Sage | v0.13.1 |
Closed | PRIDE project PXD000498, LC-MS/MS run of an unenriched Escherichia coli sample. Closed search with carbamidomethylation as fixed; oxidation as variable. | ||
| MaxQuant | v2.4.13.0 |
Closed | PRIDE project PXD025088, LC-MS/MS run of an unenriched Escherichia coli sample. Closed search with carbamidomethylation as fixed; oxidation and protein n-terminal acetylation as variable. | Input file filtered for modified peptides. | |
| Spectronaut | v9 |
Not specified | MassIVE project MSV000091214, LC-MS/MS run of an succinyl-enriched mouse kidney sample using DC8 and DC12. | ||
| PeptideAtlas | Open+Closed | PeptideAtlas protein entry P0AA16, large-scale reprocessing of public Escherichia coli data with open and closed search. | |||
| mzIdentML | Closed | PRIDE project PXD001077, LC-MS/MS of an unenriched Pyrococcus furiosus sample. PTM analysis carried out with MS-GF+ v10024 via SearchGUI v1.18.7. Closed search with carbamidomethylation as fixed; oxidation and phosphorylation as variable. | |||
| Plain/CSV | Open | IEEE VIS 2022 Bio+MedVis Challenge dataset comprising modifications of three human proteins and their mouse orthologs. PTM analysis carried out with ionbot (open search). | UniMod classes Artefact and Chemical derivative excluded (via PTMVision). |
Overview PTMs
- Shared PTM Sites between Modification Types: Shows for each modification pair, if both were found on common sites. The number of common sites is displayed with a color gradient, where brighter cells indicate lower numbers of shared sites.
- Mass Shift: Shows the mass shift of each modification type in a scatter plot. Hovering over a point will display the mass shift in Dalton.
- Site Count: Shows the number of sites that are modified with the respective modification type in a bar plot. Hovering over the bars will display the number of sites.
- PTM Class Counts: Shows how many times each of the UniMod classes have been assigned to a modification and site pair in a bar plot.
The following controls are available for the dashboard:
Screenshot of the current view.
Reset the zoom level of all overview visualizations to default (zoomed out completely).
Highlight a specific PTM of interest.
Sort the Shared PTM sites, Mass Shift and Site Count visualizations either by mass shift or count.
For more information, see the Usage Guide.
Select Single Protein of Interest
Overview of all proteins available from session data, listing one protein per row with basic information about the protein and associated PTMs. Select a single row by clicking on it (highlighted in red) and click Display to obtain a detailed view of the PTMs on the selected protein. Click symbols for help.
Filter for modifications by UniMod name
Filter for protein names/UniProt identifier
Explore Detail No Protein Selected
PTMVision offers two distinct modes to display the PTM landscape of a single protein; The Modifications View (default) and the Structure View.
The Modifications View dashboard displays:
- Per Position PTMs and Annotations: (Top) The bar plot shows the total number of modifications per position. (Mid) The presence/absence matrix, where each row represents a modification type and each column a position in the protein sequence, indicates which modification type was found at the respective position. (Bottom) Displays UniProt annotation tracks.
- Per Aminoacid PTM Counts: Displays the distribution of modification types across the different amino acid residues in a heatmap.
- Per PTM Class Counts: Shows the total number of modification sites per modification type in a bar plot.
- Legend: Coloring of all plots, excpet the Per Aminoacid PTM Counts, is based on the UniMod class of the modification. Items can be clicked to toggle visibility of the respective PTMs.
The Structure View dashboard displays:
- Per Position PTM Counts, Residue Contacts and Annotations: (Top) The bar plot shows the total number of modifications per position. (Mid) The residues are plotted against each other in a contact map; The cell color indicates whether the residues in contact are modified. Clicking on a cell highlights the respective residue in the protein structure. (Bottom) Displays UniProt annotation tracks.
- Protein Structure and Residue Contact PTM Details: Displays the 3D structure model of the protein as well as details on the obersved PTMs and distances of selected residues.
- Legend: Coloring indicates if residues in contact are modified or not as well as highlighted PTMs. Items can be clicked to toggle visibility of the respective data points.
The following controls are available for the dashboard:
Screenshot of the current view.
Reset the zoom level of all overview visualizations to default (zoomed out completely).
Highlight a specific PTM of interest.
Switch between Modifications View and Structure View.
For more information, see the Usage Guide.
